library(readr)
LOS_data <- read_csv("data/LOS.csv")Examples
Data
Length of Stay Model Data
The LOS dataset is a simulated hospital length-of-stay dataset, available through the {NHSRdatasets} R package. The dataset is licensed under creativecommons.org/publicdomain/zero/1.0.
Download CSV: LOS.csv
Download Excel: LOS.xlsx
If you have downloaded the data, and stored it in your R project folder in a folder called data, then you can read the data in:
Or if you’re using the Excel version:
library(readxl)Warning: package 'readxl' was built under R version 4.4.3LOS_data <- read_xlsx("data/LOS.xlsx")Examples
This section includes code for the examples shown. These may differ slightly from the examples shown in the live demonstration.
Example 1: Performing operations in R
See example
Make a vector using the c() function and assign it to a variable called x:
x <- c("January", "February", "March")
x[1] "January" "February" "March" There are three types of data, and we can’t mix and match them in a vector:
x <- c(23, "Yes", TRUE, 44)
x[1] "23" "Yes" "TRUE" "44" Note: you don’t get an error, but the vector probably isn’t what you expected it to be. Hint: "" means character i.e. a word.
Functions take an input(s) and produce an output. Write the function name, followed by round brackets, with the input inside the brackets. For example, calculate the exponential of 5:
exp(5)[1] 148.4132Install packages (only need to do this once):
install.packages("readxl")Load the package (each time you open R and want to use the package):
library(readxl)Example 2: Loading data into R
See example
Reading in a CSV file using base R:
LOS_data <- read.csv("data/LOS.csv")Reading in a CSV file using the {readr} package:
library(readr)
LOS_data <- read_csv("data/LOS.csv")The difference between the two is mainly the way that column names are processed.
Reading in an Excel file using the {readxl} package:
library(readxl)
LOS_data <- read_xlsx("data/LOS.xlsx")Look at the data:
View(LOS_data)Number of rows and columns
nrow(LOS_data)[1] 300ncol(LOS_data)[1] 5Example 3: Plotting single variables
See example
Histogram using the geom_histogram() function:
library(ggplot2)
ggplot(data = LOS_data, mapping = aes(x = Age)) +
geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.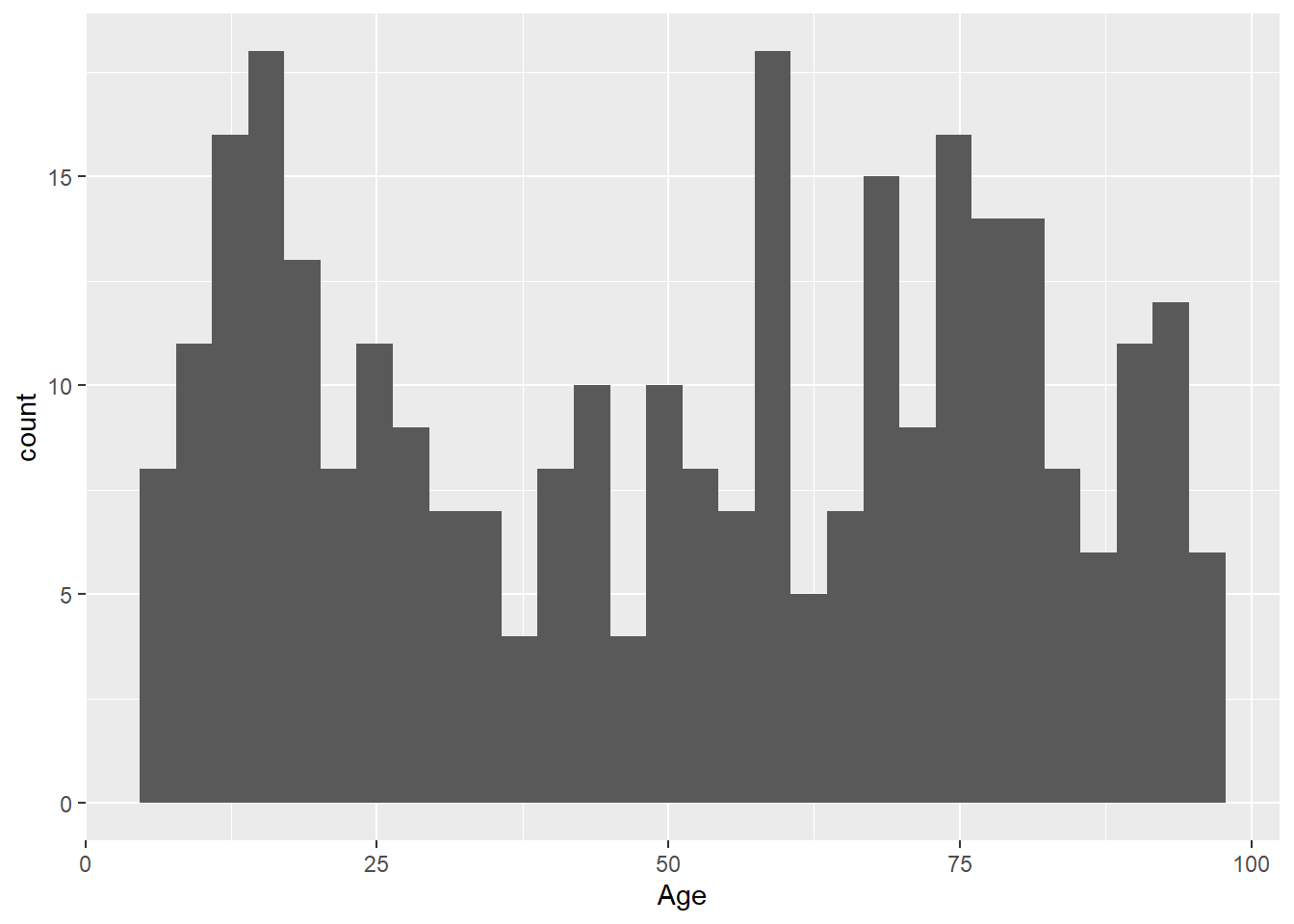
You don’t need to explicitly write the arguments:
ggplot(LOS_data, aes(x = Age)) +
geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.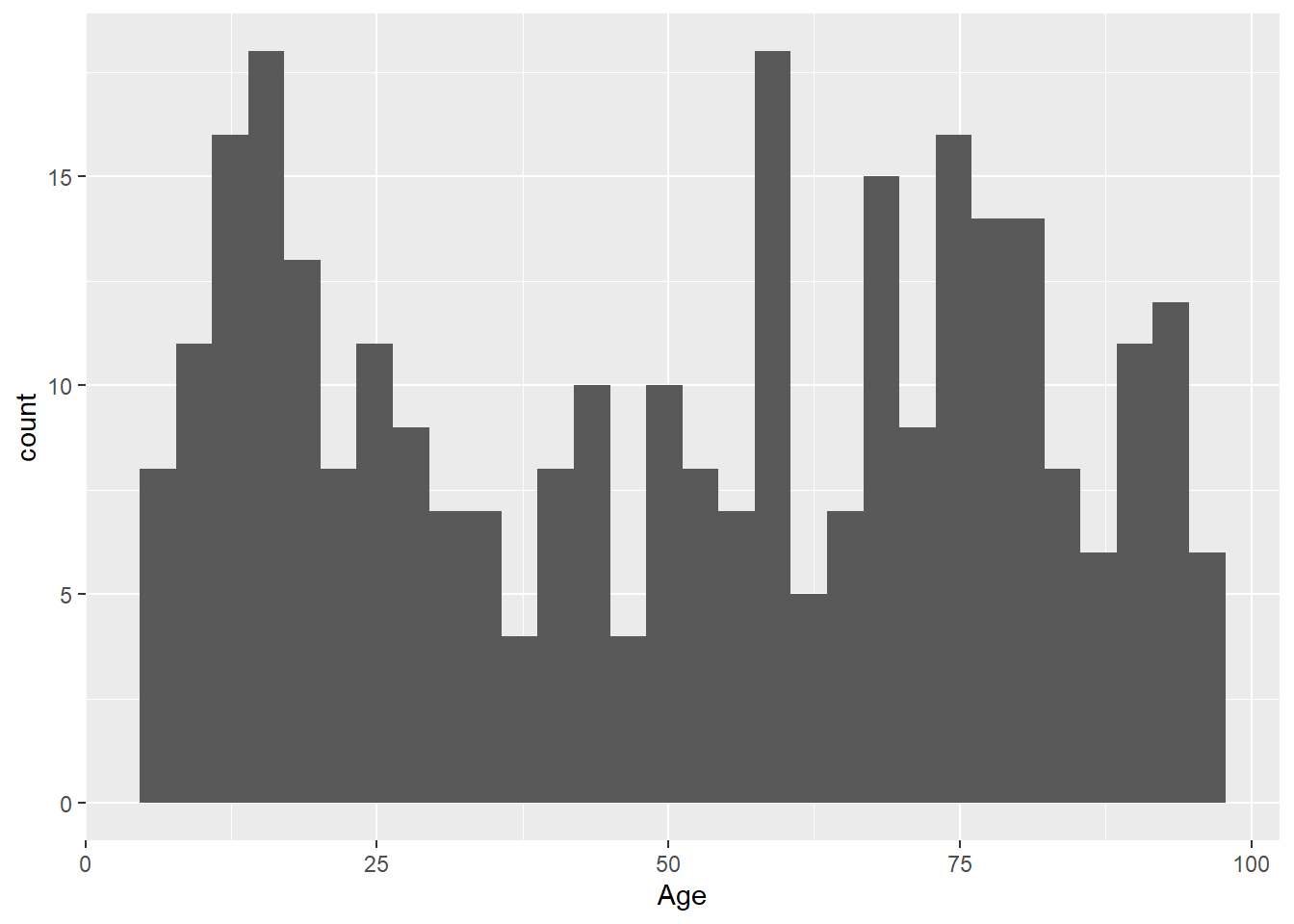
Change the binwidth:
ggplot(data = LOS_data, mapping = aes(x = Age)) +
geom_histogram(binwidth = 5)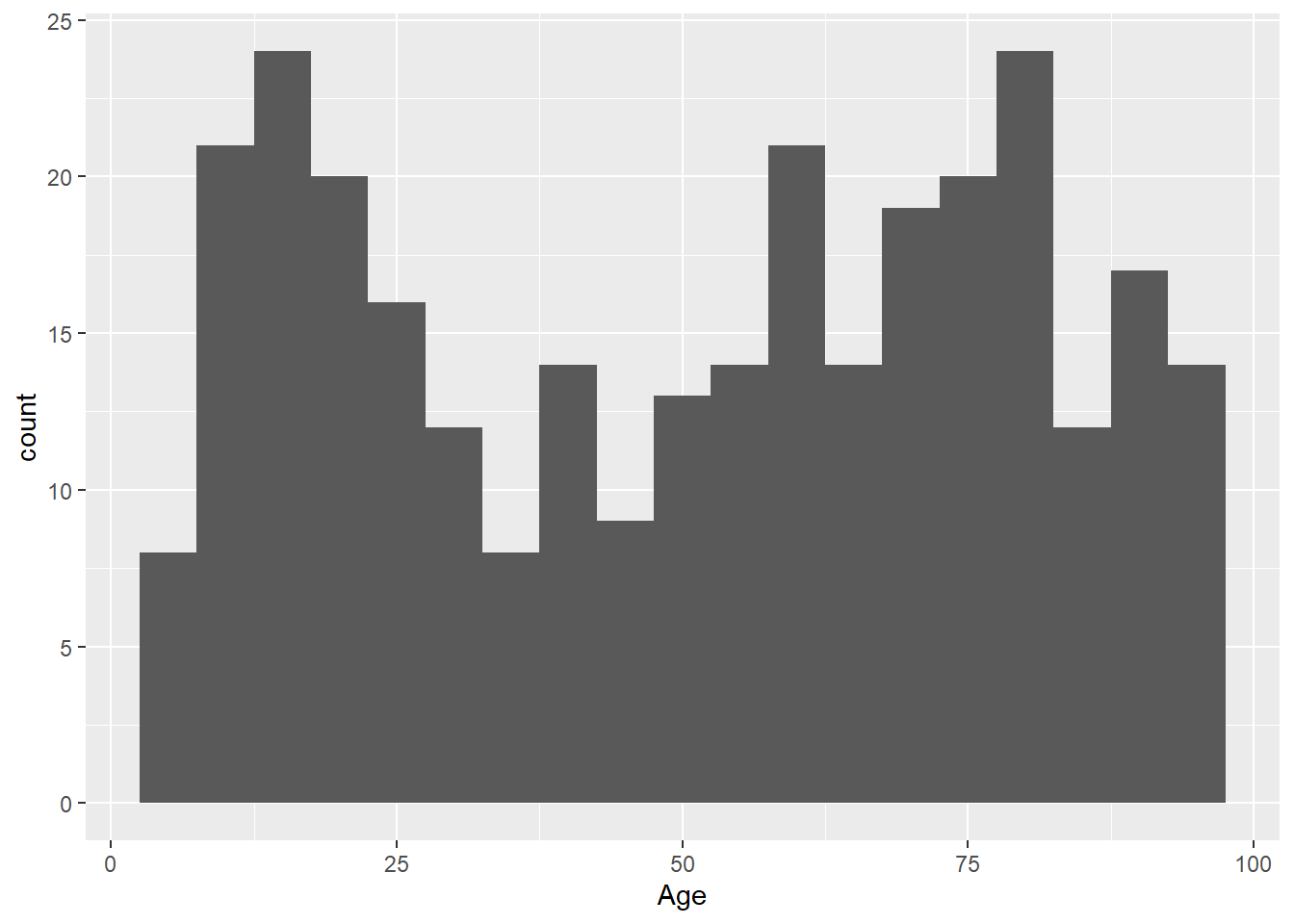
We want to subset the data to include only rows where the Organisation is equal to Trust1. We use the filter() function from the {dplyr} package to make a new data set:
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionLOS_trust1 <- LOS_data |>
filter(Organisation == "Trust1")Then use the same code to plot a histogram of age for only Trust 1 patient, but change the data we pass in:
ggplot(data = LOS_trust1, mapping = aes(x = Age)) +
geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.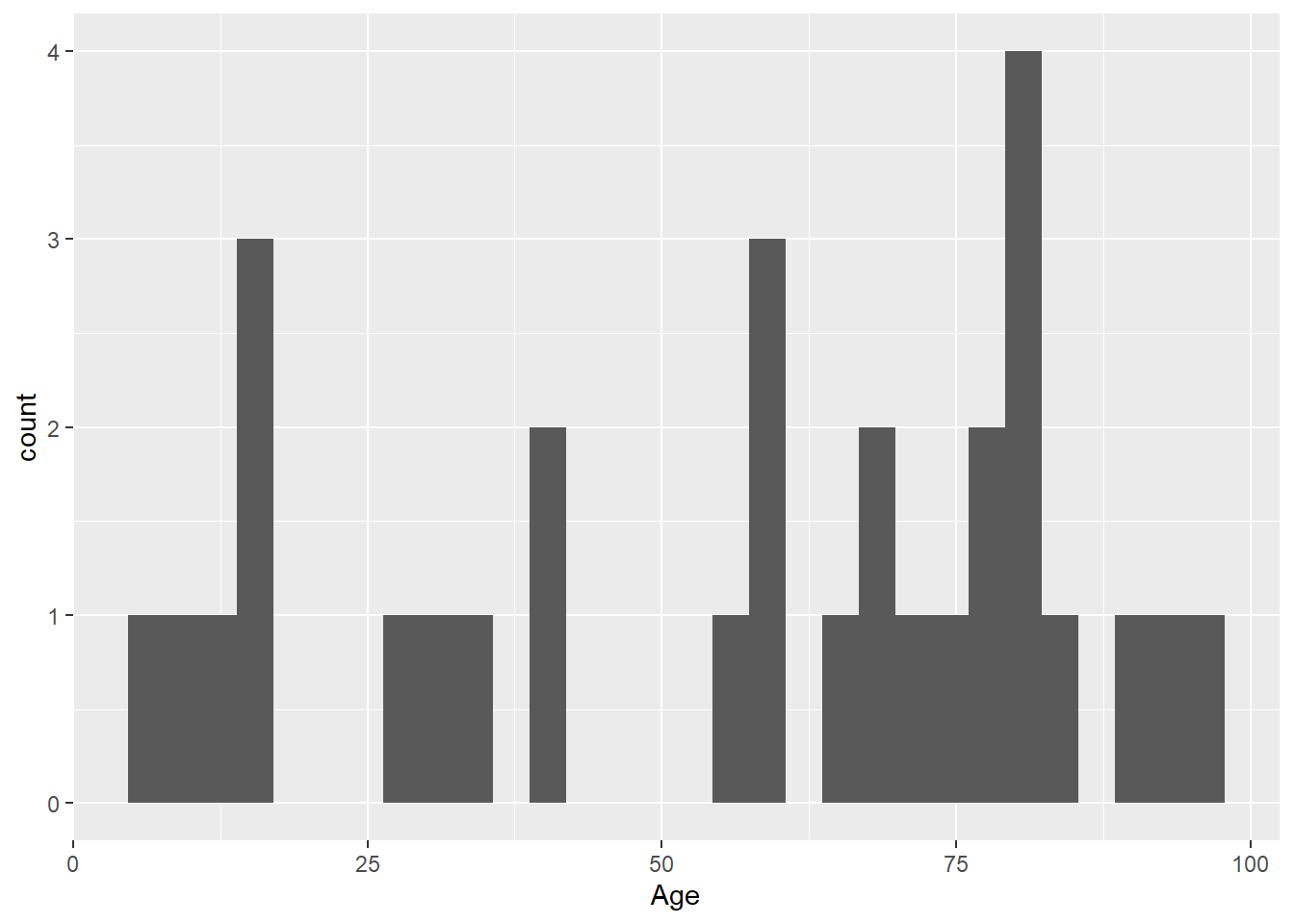
Example 4: Reading help files in R
See example
To read the help files for an R package or function, use a ? followed by the function name (with or without brackets). For example, to read the help files for the mutate() function in {dplyr}:
?mutate
?mutate()
help("mutate")Example 5: Plotting multiple variables
See example
Do older patients stay longer?
ggplot(
data = LOS_data,
mapping = aes(x = Age, y = LOS)
) +
geom_point()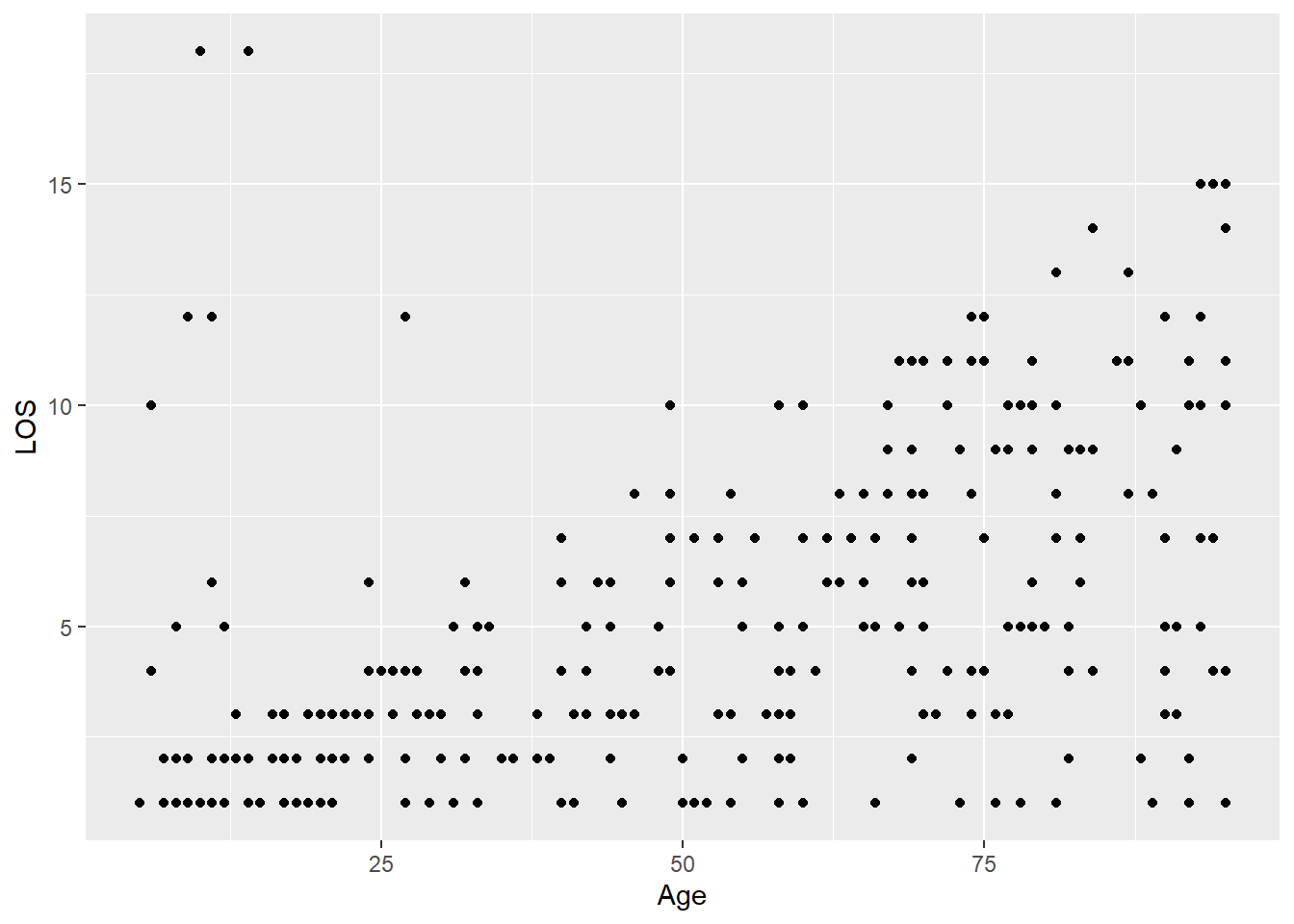
Does it vary by organisation? Let’s use colours to find out. R orders categories alphabetically unless you tell it otherwise, this is why Trust10 is before Trust2.
ggplot(
data = LOS_data,
mapping = aes(x = Age, y = LOS, colour = Organisation)
) +
geom_point()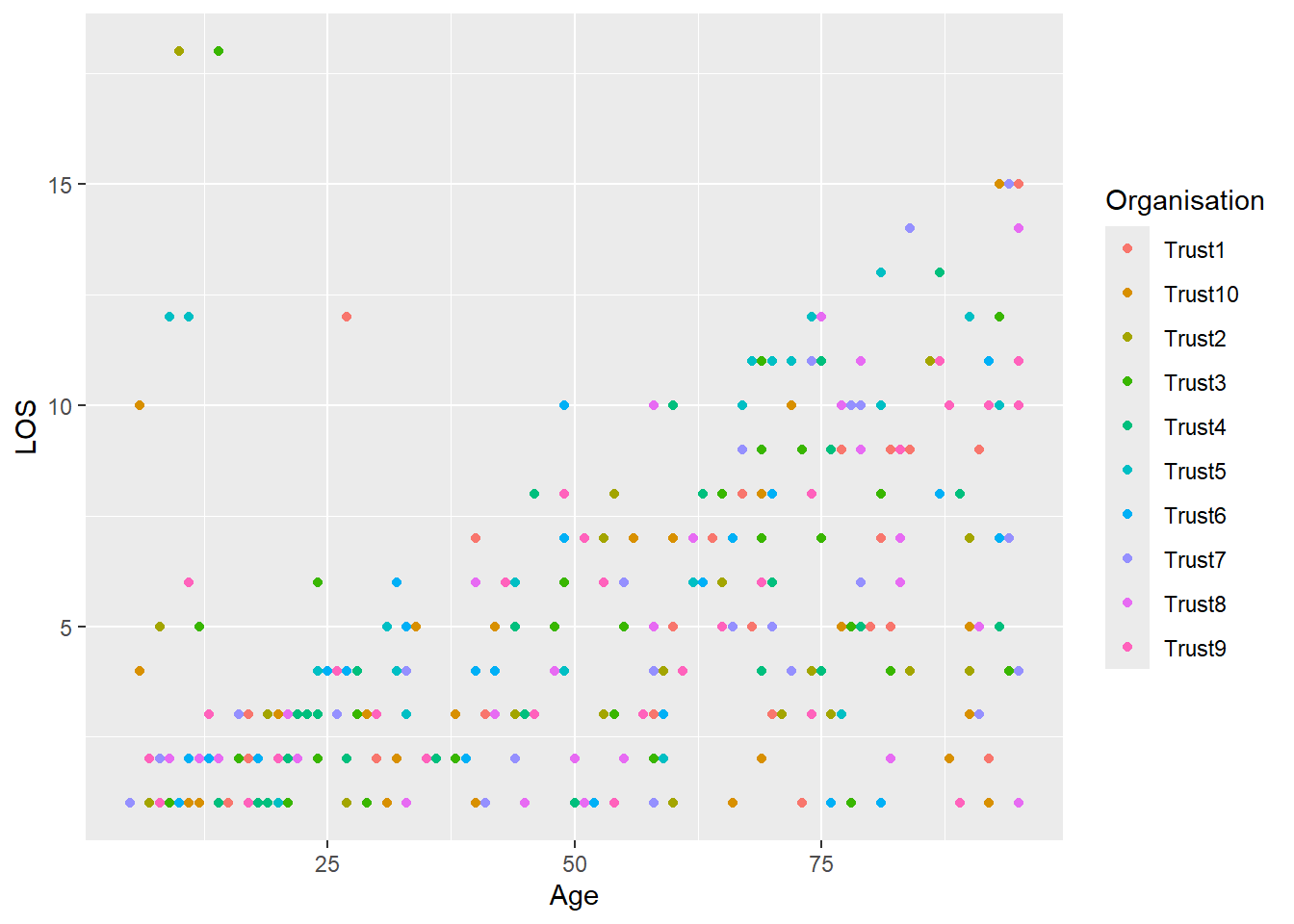
For colours based on numeric variables, R adds a continuous colour scale:
ggplot(
data = LOS_data,
mapping = aes(x = Age, y = LOS, colour = Age)
) +
geom_point()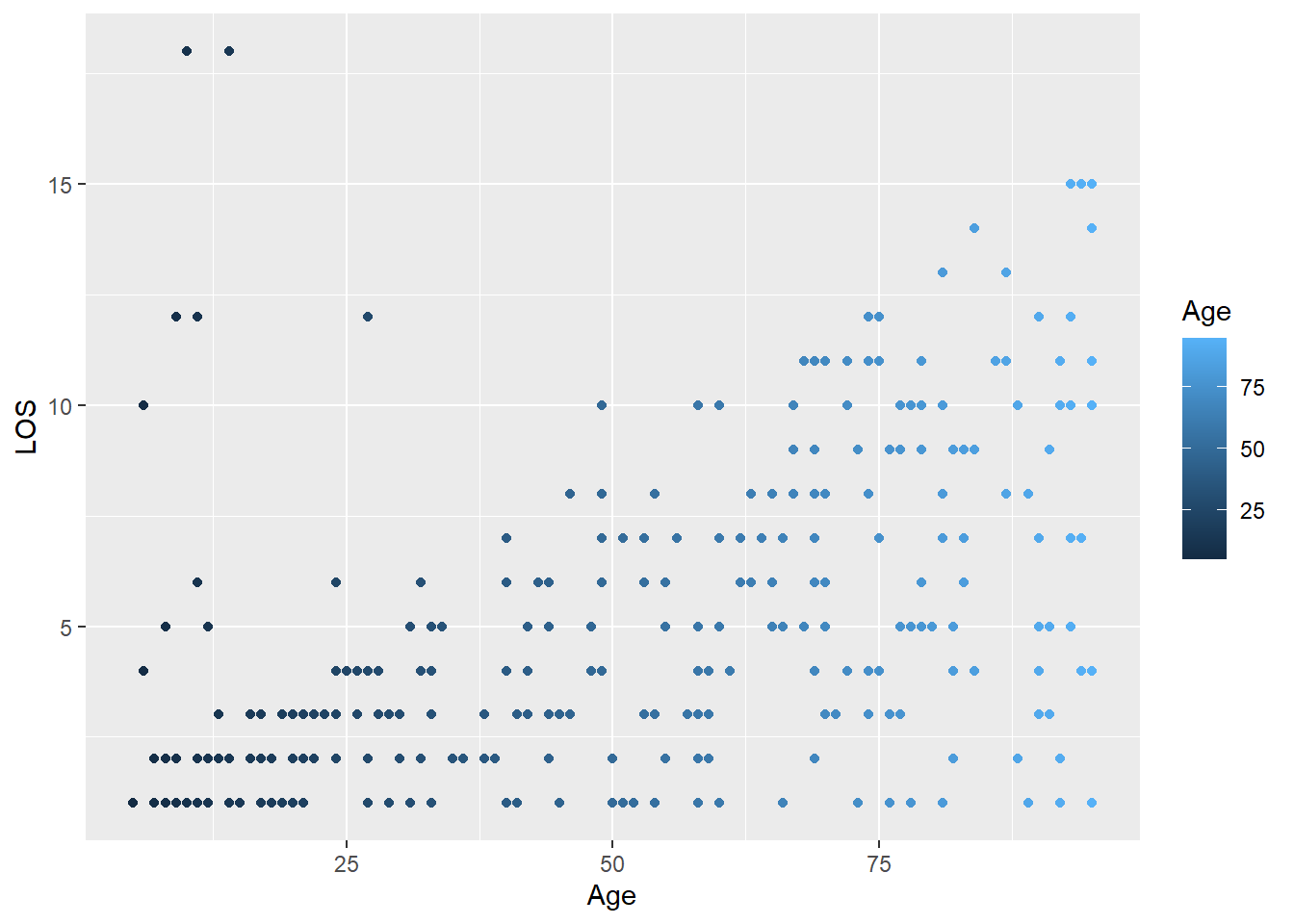
Sometimes we need to change the type of a variable for plotting e.g. from a numeric to a character.
ggplot(
data = LOS_data,
mapping = aes(x = Age, y = LOS, colour = Death)
) +
geom_point()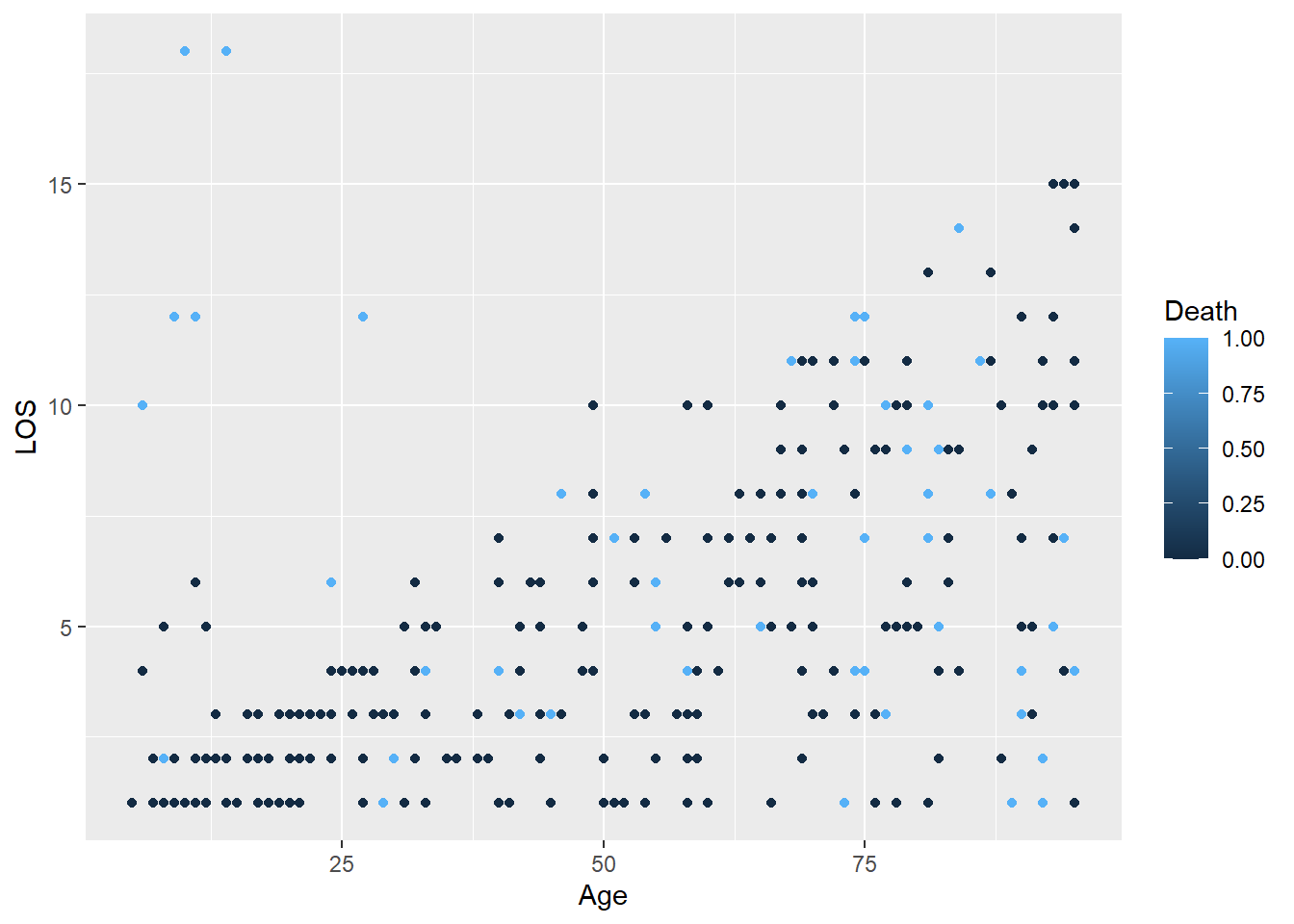
The Death column is encoded as 0 and 1 which R interprets as numbers, but these are actually categories (No and Yes). We can change it to a character or factor (ordered category).
ggplot(
data = LOS_data,
mapping = aes(x = Age, y = LOS, colour = factor(Death))
) +
geom_point()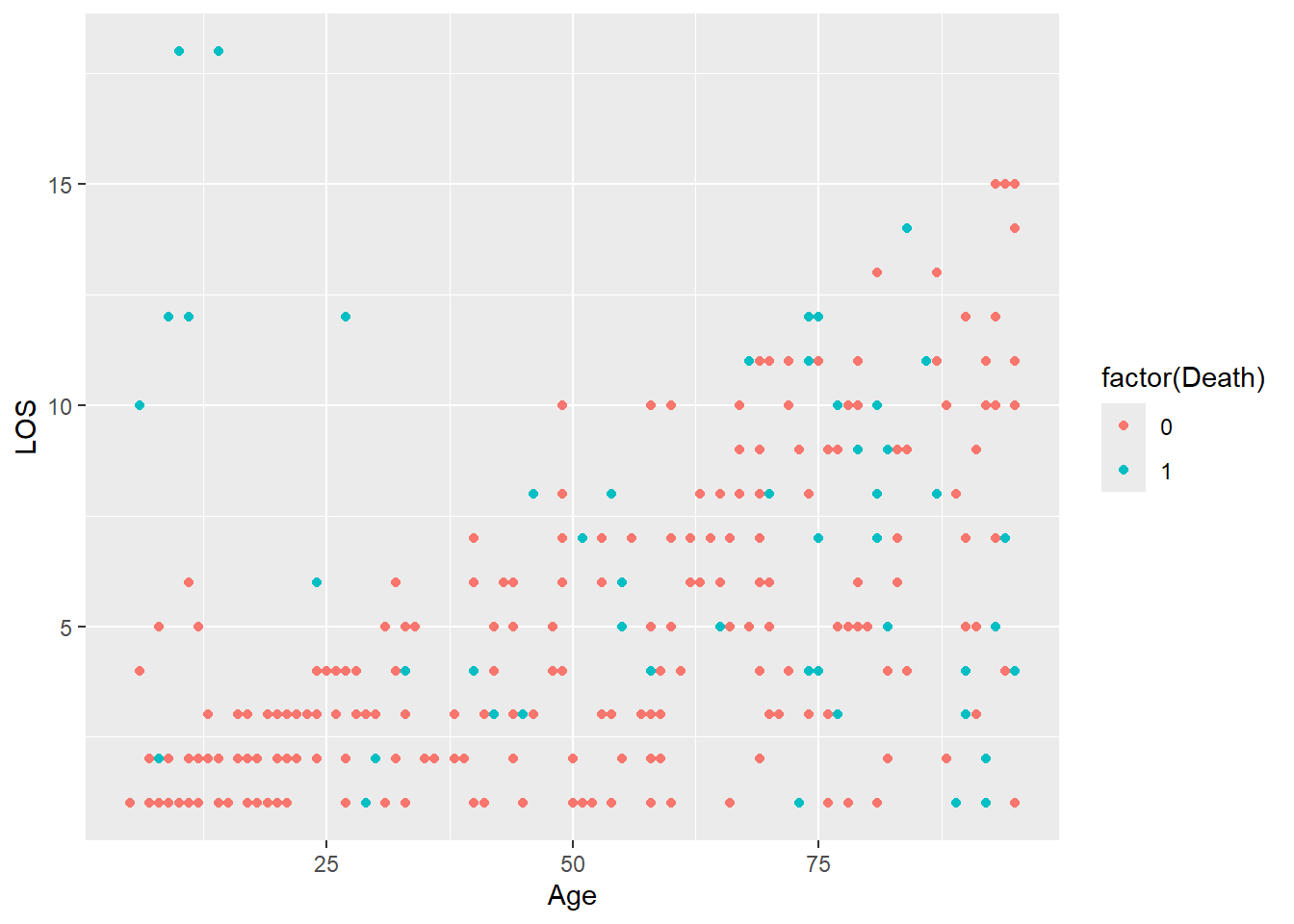
Example 6: Computing summary statistics
See example
What is the mean LOS?
In base R:
mean(LOS_data$LOS)[1] 4.936667LOS_data |>
summarise(LOS_mean = mean(LOS))# A tibble: 1 × 1
LOS_mean
<dbl>
1 4.94Calculate the average LOS for each Organisation.
LOS_data |>
group_by(Organisation) |>
summarise(LOS_mean = mean(LOS))# A tibble: 10 × 2
Organisation LOS_mean
<chr> <dbl>
1 Trust1 5.07
2 Trust10 4.3
3 Trust2 4.23
4 Trust3 5.07
5 Trust4 4.87
6 Trust5 6.1
7 Trust6 4.9
8 Trust7 5.1
9 Trust8 4.7
10 Trust9 5.03Also calculate the mean age for each Organisation:
LOS_data |>
group_by(Organisation) |>
summarise(
LOS_mean = mean(LOS),
Age_mean = mean(Age)
)# A tibble: 10 × 3
Organisation LOS_mean Age_mean
<chr> <dbl> <dbl>
1 Trust1 5.07 55.4
2 Trust10 4.3 51.0
3 Trust2 4.23 51.2
4 Trust3 5.07 47.9
5 Trust4 4.87 49.5
6 Trust5 6.1 45.7
7 Trust6 4.9 48.6
8 Trust7 5.1 53.7
9 Trust8 4.7 51.4
10 Trust9 5.03 52.3What about the standard deviation?
LOS_data |>
group_by(Organisation) |>
summarise(
LOS_mean = mean(LOS),
Age_mean = mean(Age),
LOS_sd = sd(LOS),
Age_sd = sd(Age)
)# A tibble: 10 × 5
Organisation LOS_mean Age_mean LOS_sd Age_sd
<chr> <dbl> <dbl> <dbl> <dbl>
1 Trust1 5.07 55.4 3.52 28.2
2 Trust10 4.3 51.0 3.35 29.5
3 Trust2 4.23 51.2 3.51 27.3
4 Trust3 5.07 47.9 3.98 28.4
5 Trust4 4.87 49.5 3.42 26.4
6 Trust5 6.1 45.7 4.30 28.3
7 Trust6 4.9 48.6 3.34 27.8
8 Trust7 5.1 53.7 3.82 29.2
9 Trust8 4.7 51.4 3.71 27.8
10 Trust9 5.03 52.3 3.32 28.7Example 7: Summary tables
See example
Drop the ID column, then make a summary table of the rest of the variables:
library(gtsummary)Warning: package 'gtsummary' was built under R version 4.4.3tbl1_data <- LOS_data |>
select(-ID)
tbl1_data |>
tbl_summary()| Characteristic | N = 3001 |
|---|---|
| Organisation | |
| Trust1 | 30 (10%) |
| Trust10 | 30 (10%) |
| Trust2 | 30 (10%) |
| Trust3 | 30 (10%) |
| Trust4 | 30 (10%) |
| Trust5 | 30 (10%) |
| Trust6 | 30 (10%) |
| Trust7 | 30 (10%) |
| Trust8 | 30 (10%) |
| Trust9 | 30 (10%) |
| Age | 54 (24, 76) |
| LOS | 4.0 (2.0, 7.0) |
| Death | 53 (18%) |
| 1 n (%); Median (Q1, Q3) | |
Group by Organisation:
tbl1 <- tbl1_data |>
tbl_summary(by = Organisation)
tbl1 | Characteristic | Trust1 N = 301 |
Trust10 N = 301 |
Trust2 N = 301 |
Trust3 N = 301 |
Trust4 N = 301 |
Trust5 N = 301 |
Trust6 N = 301 |
Trust7 N = 301 |
Trust8 N = 301 |
Trust9 N = 301 |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 62 (30, 80) | 49 (26, 77) | 57 (19, 74) | 52 (21, 73) | 48 (23, 75) | 39 (18, 72) | 49 (25, 74) | 58 (26, 78) | 51 (26, 79) | 54 (26, 74) |
| LOS | 5.0 (2.0, 7.0) | 3.0 (2.0, 5.0) | 3.0 (2.0, 6.0) | 4.5 (2.0, 7.0) | 4.0 (2.0, 8.0) | 4.5 (2.0, 11.0) | 4.0 (2.0, 7.0) | 4.0 (2.0, 7.0) | 3.0 (2.0, 7.0) | 4.0 (2.0, 8.0) |
| Death | 7 (23%) | 4 (13%) | 5 (17%) | 6 (20%) | 4 (13%) | 7 (23%) | 4 (13%) | 8 (27%) | 5 (17%) | 3 (10%) |
| 1 Median (Q1, Q3); n (%) | ||||||||||
Export to a Word document:
library(gt)
tbl1 |>
as_gt() |>
gtsave(filename = "Table 1.docx")Example 8: Statistical tests
See example
Is the LOS significantly different for patients under the age of 50, compare to those who are 50 or older?
# Subset data
LOS_younger <- filter(LOS_data, Age < 50)
LOS_older <- filter(LOS_data, Age >= 50)Are the means different?
t.test(LOS_younger$LOS, LOS_older$LOS)
Welch Two Sample t-test
data: LOS_younger$LOS and LOS_older$LOS
t = -8.0643, df = 296.03, p-value = 1.844e-14
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.767660 -2.289483
sample estimates:
mean of x mean of y
3.321429 6.350000 Are the variances different?
var.test(LOS_younger$LOS, LOS_older$LOS)
F test to compare two variances
data: LOS_younger$LOS and LOS_older$LOS
F = 0.64874, num df = 139, denom df = 159, p-value = 0.009204
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.4704584 0.8977698
sample estimates:
ratio of variances
0.6487431