Initial R Script
The original messy R script used for the exercises can be downloaded below.
Download .R Script: messy_exercises_script.R
See initial R script
install.packages ("readr" )install.packages ("writexl" )library (readxl)library (readexcel)library (readr)<- read_csv ("C:/Users/username/OneDrive/R code/hypoxia.csv" )View (df)# histograms hist (df$ Age) # don't use this hist (df$ BMI)hist (df$ Sleeptime)hist (df$ AHI)# An AHI of 5-14 is mild; 15-29 is moderate and 30 or more events per hour characterizes severe sleep apnea. # All normal???? # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) <- function () {<- df |> mutate (Sex = if_else (Female == 1 , "Female" , "Male" ), .after = Female) |> mutate (AHI = factor (AHI))return (df)<- myfunction ()library (ggplot2)ggplot (df) + geom_histogram (aes (Age))ggplot (df) + geom_histogram (aes (AHI))ggplot (df) + geom_bar (aes (AHI))library (ggcorrplot)# doesn't work error # ggcorrplot::ggcorrplot(cor(df)) |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()# save as plot - PNG format # width 5 inch, height = 5 inch for paper ggplot (df) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (df) + geom_bar (aes (AHI)) + facet_wrap (Race~ Sex) ggplot (df) + geom_bar (aes (AHI)) + facet_grid (Race~ Sex) ggplot (df) + geom_bar (aes (AHI, fill = Diabetes)) # no colours <- df |> mutate (Diabetesfct = factor (Diabetes))ggplot (df) + geom_bar (aes (AHI, fill= Diabetesfct),position = "fill" )+ scale_fill_brewer (palette = "Dark2" )+ theme_minimal () # no colours library (dplyr)mean (df$ Age) #50.65667 sd (df$ Age)mean (df$ BMI) #46.74875 table (df$ AHI) #?missing table (df$ ` Duration of Surg ` )mean (df$ ` Duration of Surg ` ) #4.314947 # error - don't know why #tests chisq.test (table (df$ AHI, df$ Sex))chisq.test (table (df$ AHI, df$ Race))chisq.test (table (df$ AHI, df$ Smoking))chisq.test (table (df$ AHI, df$ Diabetes))<- df |> filter (BMI <= median (BMI))<- df |> filter (BMI > median (BMI))t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)$ p.value# models <- df |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 ))<- glm (severeAHI~ ., data = df, family = "binomial" )summary (mod1)<- df |> select (- c (Female, AHI))<- glm (severe_AHI~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI~ Age+ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI~ Age+ Sex, data= mod_data, family= "binomial" )summary (mod3)<- glm (severe_AHI~ Age+ BMI, data= mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI~ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI~ Age, data= mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI~ Sex, data= mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI~ BMI, data= mod_data, family = "binomial" )summary (mod8)c (mod2$ aic, mod3$ aic, mod4$ aic, mod5$ aic, mod6$ aic, mod7$ aic, mod8$ aic)# mod2 lowest # results library (ggforestplot)library (forestmodel)forest_model (mod2)ggsave ("forestplot.png" )# saved as forestplot.png
Exercises
Exercise 1: Projects and relative file paths
Create an R project for today’s workshop - name it something sensible!
Download the messy_exercises_script.R script from nrennie.rbind.io/training-better-r-code/exercises.html and the hypoxia.csv data.
Add those files to your R project in a sensible way.
Edit the Global / Project options appropriately.
Edit the script to use relative file paths.
Solution
Folder structure:
R script:
install.packages ("readr" )install.packages ("writexl" )library (readxl)library (readexcel)library (readr)<- read_csv ("hypoxia.csv" )View (df)# histograms hist (df$ Age) # don't use this hist (df$ BMI)hist (df$ Sleeptime)hist (df$ AHI)# An AHI of 5-14 is mild; 15-29 is moderate and 30 or more events per hour characterizes severe sleep apnea. # All normal???? # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) <- function () {<- df |> mutate (Sex = if_else (Female == 1 , "Female" , "Male" ), .after = Female) |> mutate (AHI = factor (AHI))return (df)<- myfunction ()library (ggplot2)ggplot (df) + geom_histogram (aes (Age))ggplot (df) + geom_histogram (aes (AHI))ggplot (df) + geom_bar (aes (AHI))library (ggcorrplot)# doesn't work error # ggcorrplot::ggcorrplot(cor(df)) |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()# save as plot - PNG format # width 5 inch, height = 5 inch for paper ggplot (df) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (df) + geom_bar (aes (AHI)) + facet_wrap (Race~ Sex) ggplot (df) + geom_bar (aes (AHI)) + facet_grid (Race~ Sex) ggplot (df) + geom_bar (aes (AHI, fill = Diabetes)) # no colours <- df |> mutate (Diabetesfct = factor (Diabetes))ggplot (df) + geom_bar (aes (AHI, fill= Diabetesfct),position = "fill" )+ scale_fill_brewer (palette = "Dark2" )+ theme_minimal () # no colours library (dplyr)mean (df$ Age) #50.65667 sd (df$ Age)mean (df$ BMI) #46.74875 table (df$ AHI) #?missing table (df$ ` Duration of Surg ` )mean (df$ ` Duration of Surg ` ) #4.314947 # error - don't know why #tests chisq.test (table (df$ AHI, df$ Sex))chisq.test (table (df$ AHI, df$ Race))chisq.test (table (df$ AHI, df$ Smoking))chisq.test (table (df$ AHI, df$ Diabetes))<- df |> filter (BMI <= median (BMI))<- df |> filter (BMI > median (BMI))t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)$ p.value# models <- df |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 ))<- glm (severeAHI~ ., data = df, family = "binomial" )summary (mod1)<- df |> select (- c (Female, AHI))<- glm (severe_AHI~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI~ Age+ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI~ Age+ Sex, data= mod_data, family= "binomial" )summary (mod3)<- glm (severe_AHI~ Age+ BMI, data= mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI~ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI~ Age, data= mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI~ Sex, data= mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI~ BMI, data= mod_data, family = "binomial" )summary (mod8)c (mod2$ aic, mod3$ aic, mod4$ aic, mod5$ aic, mod6$ aic, mod7$ aic, mod8$ aic)# mod2 lowest # results library (ggforestplot)library (forestmodel)forest_model (mod2)ggsave ("forestplot.png" )# saved as forestplot.png
Exercise 2: Organising a script
Reorganise the messy R script by adding sections and subsections.
Edit the comments in the document to make them more useful.
Rename variables and functions if you think they need to be renamed.
Solution
# Load packages ----------------------------------------------------------- library (readr)library (ggplot2)library (dplyr)library (ggforestplot)library (forestmodel)library (ggcorrplot)# Read data --------------------------------------------------------------- <- read_csv ("data/hypoxia.csv" )View (hypoxia)# Useful functions -------------------------------------------------------- # An AHI of 5-14 is mild; 15-29 is moderate and # 30 or more events per hour characterizes severe sleep apnea. # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) # Convert binary `Female` column to characters for Male and Female <- function (data = hypoxia) {<- data |> mutate (Sex = if_else (Female == 1 , "Female" , "Male" ), .after = Female) |> mutate (AHI = factor (AHI))return (output)<- convert_to_factors ()# Exploratory plots ------------------------------------------------------- # Histograms and bar charts ggplot (hypoxia) + geom_histogram (aes (Age))ggplot (hypoxia) + geom_histogram (aes (AHI))ggplot (hypoxia) + geom_bar (aes (AHI))# Correlation |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()ggsave ("corr_plot.png" , width = 5 , height = 5 )# Multiple factors ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (Race~ Sex) ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_grid (Race~ Sex) <- hypoxia |> mutate (Diabetesfct = factor (Diabetes))ggplot (hypoxia) + geom_bar (aes (AHI, fill= Diabetesfct),position = "fill" )+ scale_fill_brewer (palette = "Dark2" )+ theme_minimal () # no colours # Summary statistics ------------------------------------------------------ # Mean mean (hypoxia$ Age)mean (hypoxia$ BMI)mean (hypoxia$ ` Duration of Surg ` )# Standard Deviation sd (hypoxia$ Age)# Counts table (hypoxia$ AHI)table (hypoxia$ ` Duration of Surg ` )# Statistical tests ------------------------------------------------------- # Chi-squared tests of AHI factors chisq.test (table (hypoxia$ AHI, hypoxia$ Sex))chisq.test (table (hypoxia$ AHI, hypoxia$ Race))chisq.test (table (hypoxia$ AHI, hypoxia$ Smoking))chisq.test (table (hypoxia$ AHI, hypoxia$ Diabetes))# T-tests for BMI and Sleep time # Assume variance equal <- hypoxia |> filter (BMI <= median (BMI))<- hypoxia |> filter (BMI > median (BMI))t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)$ p.value# Modelling --------------------------------------------------------------- <- hypoxia |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 )) |> select (- c (Female, AHI))<- glm (severe_AHI~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI~ Age+ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI~ Age+ Sex, data= mod_data, family= "binomial" )summary (mod3)<- glm (severe_AHI~ Age+ BMI, data= mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI~ Sex+ BMI, data= mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI~ Age, data= mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI~ Sex, data= mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI~ BMI, data= mod_data, family = "binomial" )summary (mod8)# Results ----------------------------------------------------------------- # AIC <- c (mod2$ aic, mod3$ aic, mod4$ aic, mod5$ aic, mod6$ aic, mod7$ aic, mod8$ aic)# Forest plot of best model forest_model (mod2)ggsave ("forestplot.png" )
Exercise 3: Styling scripts
Install and load the lintr and styler packages if you don’t already use them.
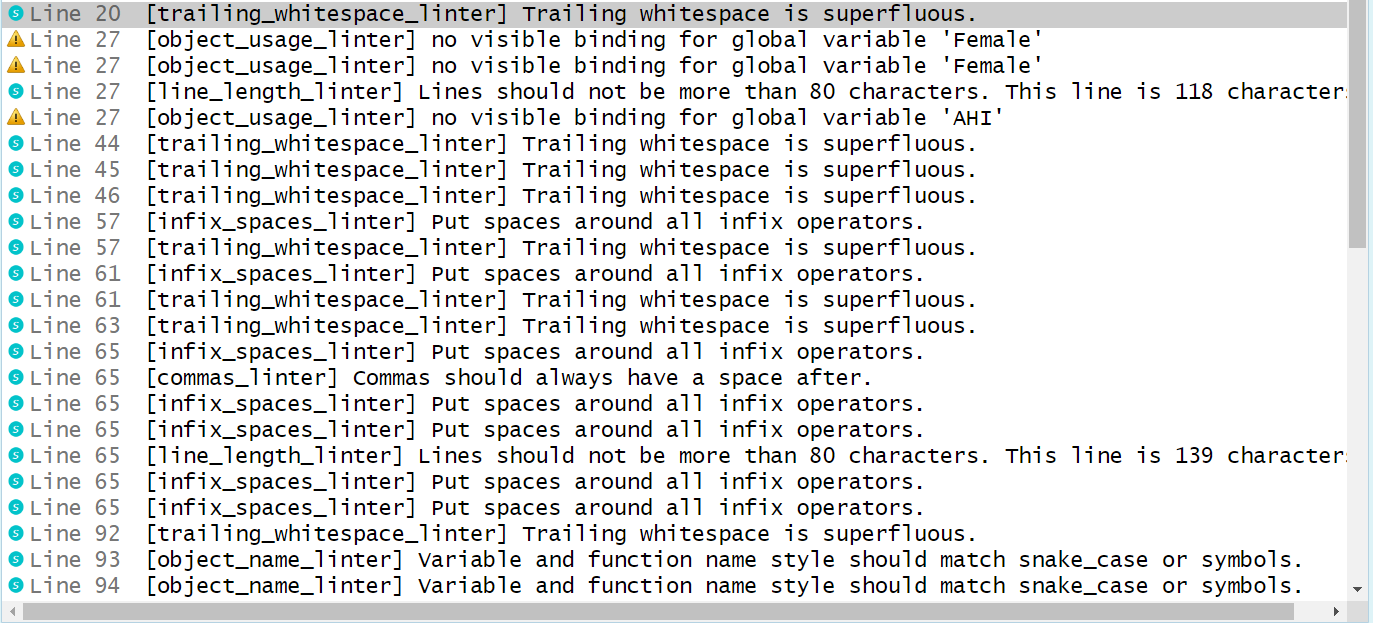
Run lint() on the messy R script. Do you understand all of the messages?
Run style_file(). What has changed in your script?
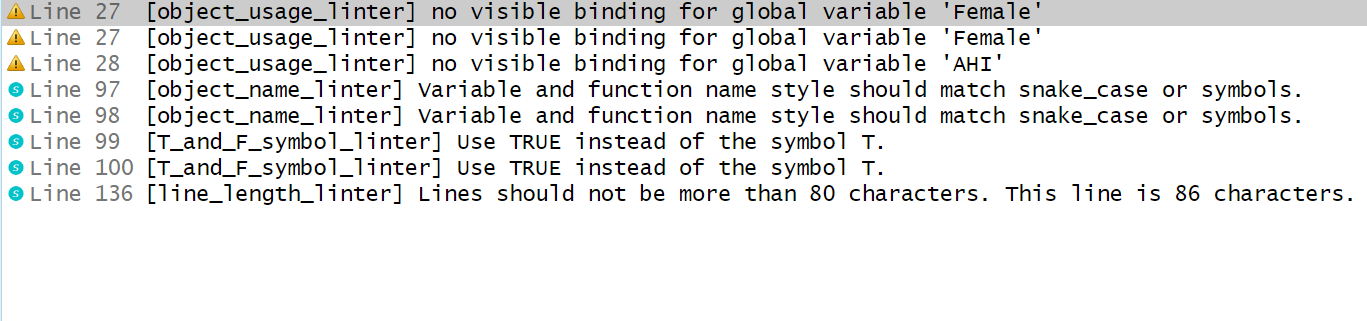
Re-run lint() on the script. Have all of the issues been fixed? If not, manually implement changes to the file.
Bonus : Add an RStudio keyboard shortcut for style_active_file().
Solution
Running lintr::lint("messy_exercises_script.R"):
After running styler::style_active_file():
# Load packages ----------------------------------------------------------- library (readr)library (ggplot2)library (dplyr)library (ggforestplot)library (forestmodel)library (ggcorrplot)# Read data --------------------------------------------------------------- <- read_csv ("data/hypoxia.csv" )View (hypoxia)# Useful functions -------------------------------------------------------- # An AHI of 5-14 is mild; 15-29 is moderate and # 30 or more events per hour characterizes severe sleep apnea. # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) # Convert binary `Female` column to characters for Male and Female <- function (data = hypoxia) {<- data |> mutate (Sex = if_else (Female == 1 , "Female" , "Male" ), .after = Female) |> mutate (AHI = factor (AHI))return (output)<- convert_to_factors ()# Exploratory plots ------------------------------------------------------- # Histograms and bar charts ggplot (hypoxia) + geom_histogram (aes (Age))ggplot (hypoxia) + geom_histogram (aes (AHI))ggplot (hypoxia) + geom_bar (aes (AHI))# Correlation |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()ggsave ("corr_plot.png" , width = 5 , height = 5 )# Multiple factors ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (Race ~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_grid (Race ~ Sex)<- hypoxia |> mutate (Diabetesfct = factor (Diabetes))ggplot (hypoxia) + geom_bar (aes (AHI, fill = Diabetesfct), position = "fill" ) + scale_fill_brewer (palette = "Dark2" ) + theme_minimal () # no colours # Summary statistics ------------------------------------------------------ # Mean mean (hypoxia$ Age)mean (hypoxia$ BMI)mean (hypoxia$ ` Duration of Surg ` )# Standard Deviation sd (hypoxia$ Age)# Counts table (hypoxia$ AHI)table (hypoxia$ ` Duration of Surg ` )# Statistical tests ------------------------------------------------------- # Chi-squared tests of AHI factors chisq.test (table (hypoxia$ AHI, hypoxia$ Sex))chisq.test (table (hypoxia$ AHI, hypoxia$ Race))chisq.test (table (hypoxia$ AHI, hypoxia$ Smoking))chisq.test (table (hypoxia$ AHI, hypoxia$ Diabetes))# T-tests for BMI and Sleep time # Assume variance equal <- hypoxia |> filter (BMI <= median (BMI))<- hypoxia |> filter (BMI > median (BMI))t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = T)$ p.value# Modelling --------------------------------------------------------------- <- hypoxia |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 )) |> select (- c (Female, AHI))<- glm (severe_AHI ~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI ~ Age + Sex + BMI, data = mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI ~ Age + Sex, data = mod_data, family = "binomial" )summary (mod3)<- glm (severe_AHI ~ Age + BMI, data = mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI ~ Sex + BMI, data = mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI ~ Age, data = mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI ~ Sex, data = mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI ~ BMI, data = mod_data, family = "binomial" )summary (mod8)# Results ----------------------------------------------------------------- # AIC <- c (mod2$ aic, mod3$ aic, mod4$ aic, mod5$ aic, mod6$ aic, mod7$ aic, mod8$ aic)# Forest plot of best model forest_model (mod2)ggsave ("forestplot.png" )
Running lintr::lint("messy_exercises_script.R") again:
After manual tidy up:
# Load packages ----------------------------------------------------------- library (readr)library (ggplot2)library (dplyr)library (ggforestplot)library (forestmodel)library (ggcorrplot)# Read data --------------------------------------------------------------- <- read_csv ("data/hypoxia.csv" )View (hypoxia)# Useful functions -------------------------------------------------------- # An AHI of 5-14 is mild; 15-29 is moderate and # 30 or more events per hour characterizes severe sleep apnea. # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) # Convert binary `Female` column to characters for Male and Female <- function (data = hypoxia) {<- data |> mutate (Sex = if_else (.data$ Female == 1 , "Female" , "Male" ),.after = .data$ Female|> mutate (AHI = factor (.data$ AHI))return (output)<- convert_to_factors ()# Exploratory plots ------------------------------------------------------- # Histograms and bar charts ggplot (hypoxia) + geom_histogram (aes (Age))ggplot (hypoxia) + geom_histogram (aes (AHI))ggplot (hypoxia) + geom_bar (aes (AHI))# Correlation |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()ggsave ("corr_plot.png" , width = 5 , height = 5 )# Multiple factors ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (Race ~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_grid (Race ~ Sex)<- hypoxia |> mutate (Diabetesfct = factor (Diabetes))ggplot (hypoxia) + geom_bar (aes (AHI, fill = Diabetesfct), position = "fill" ) + scale_fill_brewer (palette = "Dark2" ) + theme_minimal () # no colours # Summary statistics ------------------------------------------------------ # Mean mean (hypoxia$ Age)mean (hypoxia$ BMI)mean (hypoxia$ ` Duration of Surg ` )# Standard Deviation sd (hypoxia$ Age)# Counts table (hypoxia$ AHI)table (hypoxia$ ` Duration of Surg ` )# Statistical tests ------------------------------------------------------- # Chi-squared tests of AHI factors chisq.test (table (hypoxia$ AHI, hypoxia$ Sex))chisq.test (table (hypoxia$ AHI, hypoxia$ Race))chisq.test (table (hypoxia$ AHI, hypoxia$ Smoking))chisq.test (table (hypoxia$ AHI, hypoxia$ Diabetes))# T-tests for BMI and Sleep time # Assume variance equal <- hypoxia |> filter (BMI <= median (BMI)) # nolint <- hypoxia |> filter (BMI > median (BMI)) # nolint t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = TRUE )t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = TRUE )$ p.value# Modelling --------------------------------------------------------------- <- hypoxia |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 )) |> select (- c (Female, AHI))<- glm (severe_AHI ~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI ~ Age + Sex + BMI, data = mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI ~ Age + Sex, data = mod_data, family = "binomial" )summary (mod3)<- glm (severe_AHI ~ Age + BMI, data = mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI ~ Sex + BMI, data = mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI ~ Age, data = mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI ~ Sex, data = mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI ~ BMI, data = mod_data, family = "binomial" )summary (mod8)# Results ----------------------------------------------------------------- # AIC <- c ($ aic, mod3$ aic, mod4$ aic,$ aic, mod6$ aic, mod7$ aic, mod8$ aic# Forest plot of best model forest_model (mod2)ggsave ("forestplot.png" )
Note: linters aren’t always correct - use # nolint to ignore a line.
Exercise 4: Multiple scripts and folders
Re-organise your R project folder with multiple sub-directories.
Split your messy R script into multiple files, that are appropriately named.
What is the order and dependencies of each script?
Solution
Folder structure:
# Load packages ----------------------------------------------------------- library (readr)library (ggplot2)library (dplyr)library (ggforestplot)library (forestmodel)library (ggcorrplot)# Useful functions -------------------------------------------------------- # An AHI of 5-14 is mild; 15-29 is moderate and # 30 or more events per hour characterizes severe sleep apnea. # 1 = (AHI < 5); 2 = (5 ≤ AHI < 15); # 3 = (15 ≤ AHI < 30); 4 = (AHI ≥ 30) # Convert binary `Female` column to characters for Male and Female <- function (data = hypoxia) {<- data |> mutate (Sex = if_else (.data$ Female == 1 , "Female" , "Male" ),.after = .data$ Female|> mutate (AHI = factor (.data$ AHI))return (output)
source ("R/00_packages.R" )<- read_csv ("data/hypoxia.csv" )<- convert_to_factors (hypoxia)
02_exploratory_analysis.R
source ("R/01_load_data.R" )# Exploratory plots ------------------------------------------------------- # Histograms and bar charts ggplot (hypoxia) + geom_histogram (aes (Age))ggplot (hypoxia) + geom_histogram (aes (AHI))ggplot (hypoxia) + geom_bar (aes (AHI))# Correlation |> select (where (is.numeric)) |> cor (use = "complete.obs" ) |> ggcorrplot ()ggsave ("plots/corr_plot.png" , width = 5 , height = 5 )# Multiple factors ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_wrap (Race ~ Sex)ggplot (hypoxia) + geom_bar (aes (AHI)) + facet_grid (Race ~ Sex)<- hypoxia |> mutate (Diabetesfct = factor (Diabetes))ggplot (hypoxia) + geom_bar (aes (AHI, fill = Diabetesfct), position = "fill" ) + scale_fill_brewer (palette = "Dark2" ) + theme_minimal () # no colours # Summary statistics ------------------------------------------------------ # Mean mean (hypoxia$ Age)mean (hypoxia$ BMI)mean (hypoxia$ ` Duration of Surg ` )# Standard Deviation sd (hypoxia$ Age)# Counts table (hypoxia$ AHI)table (hypoxia$ ` Duration of Surg ` )# Statistical tests ------------------------------------------------------- # Chi-squared tests of AHI factors chisq.test (table (hypoxia$ AHI, hypoxia$ Sex))chisq.test (table (hypoxia$ AHI, hypoxia$ Race))chisq.test (table (hypoxia$ AHI, hypoxia$ Smoking))chisq.test (table (hypoxia$ AHI, hypoxia$ Diabetes))# T-tests for BMI and Sleep time # Assume variance equal <- hypoxia |> filter (BMI <= median (BMI)) # nolint <- hypoxia |> filter (BMI > median (BMI)) # nolint t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = TRUE )t.test (BMI1$ Sleeptime, BMI2$ Sleeptime, var.equal = TRUE )$ p.value
source ("R/01_load_data.R" )# Modelling --------------------------------------------------------------- <- hypoxia |> mutate (severeAHI = if_else (AHI == 4 , 1 , 0 )) |> select (- c (Female, AHI))<- glm (severe_AHI ~ ., data = mod_data, family = "binomial" )summary (mod1)<- glm (severe_AHI ~ Age + Sex + BMI, data = mod_data, family = "binomial" )summary (mod2)<- glm (severe_AHI ~ Age + Sex, data = mod_data, family = "binomial" )summary (mod3)<- glm (severe_AHI ~ Age + BMI, data = mod_data, family = "binomial" )summary (mod4)<- glm (severe_AHI ~ Sex + BMI, data = mod_data, family = "binomial" )summary (mod5)<- glm (severe_AHI ~ Age, data = mod_data, family = "binomial" )summary (mod6)<- glm (severe_AHI ~ Sex, data = mod_data, family = "binomial" )summary (mod7)<- glm (severe_AHI ~ BMI, data = mod_data, family = "binomial" )summary (mod8)# Results ----------------------------------------------------------------- # AIC <- c ($ aic, mod3$ aic, mod4$ aic,$ aic, mod6$ aic, mod7$ aic, mod8$ aic# Forest plot of best model forest_model (mod2)ggsave ("plots/forestplot.png" )
Exercise 5: Other useful tips
The following code doesn’t work. Imagine you are not allowed to share the hypoxia data with anyone else. Build a reprex that you could share with someone else.
library (ggplot2)ggplot (hypoxia) + geom_col (aes (AHI, fill = Smoking))
Solution
```
library(ggplot2)
library(tibble)
hypoxia_reprex <- tribble(
~AHI, ~Smoking,
2, 0,
3, 1,
2, 0,
4, 0,
1, 0,
1, 0,
3, 0
)
ggplot(hypoxia_reprex) +
geom_col(aes(AHI, fill = Smoking))
#> Error in `geom_col()`:
#> ! Problem while setting up geom.
#> ℹ Error occurred in the 1st layer.
#> Caused by error in `compute_geom_1()`:
#> ! `geom_col()` requires the following missing aesthetics: y.
```
```
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.4.1 (2024-06-14 ucrt)
#> os Windows 11 x64 (build 22000)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United Kingdom.utf8
#> ctype English_United Kingdom.utf8
#> tz Europe/London
#> date 2024-10-21
#> pandoc 3.1.11 @ C:/Program Files/RStudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.2)
#> colorspace 2.1-1 2023-10-23 [1] R-Forge (R 4.3.1)
#> digest 0.6.35 2024-03-11 [1] CRAN (R 4.4.0)
#> dplyr 1.1.4 2023-11-17 [1] CRAN (R 4.3.2)
#> evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.2)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.2)
#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
#> fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
#> fs 1.6.4 2024-04-25 [1] CRAN (R 4.4.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
#> ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.3.3)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.2)
#> gtable 0.3.5 2024-04-22 [1] CRAN (R 4.3.3)
#> htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.4.0)
#> knitr 1.47 2024-05-29 [1] CRAN (R 4.3.3)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.2)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
#> munsell 0.5.1 2024-04-01 [1] CRAN (R 4.3.3)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
#> purrr 1.0.2 2023-08-10 [1] CRAN (R 4.3.1)
#> R.cache 0.16.0 2022-07-21 [1] CRAN (R 4.3.0)
#> R.methodsS3 1.8.2 2022-06-13 [1] CRAN (R 4.3.0)
#> R.oo 1.26.0 2024-01-24 [1] CRAN (R 4.3.2)
#> R.utils 2.12.3 2023-11-18 [1] CRAN (R 4.3.2)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
#> reprex 2.1.0 2024-01-11 [1] CRAN (R 4.3.2)
#> rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.2)
#> rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.3.3)
#> rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.3.3)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.2)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
#> styler 1.10.3 2024-04-07 [1] CRAN (R 4.3.3)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
#> tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.3)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.2)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.2)
#> withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.2)
#> xfun 0.44 2024-05-15 [1] CRAN (R 4.4.0)
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.2)
#>
#> [1] C:/Users/rennien2/AppData/Local/R/win-library/4.4
#> [2] C:/Users/rennien2/AppData/Local/Programs/R/R-4.4.1/library
#>
#> ──────────────────────────────────────────────────────────────────────────────
```